Easily Screen 2 Billion Molecules
Connect to our public HNSW service with your own scoring function and begin virtual screening of 2 billion molecules in minutes
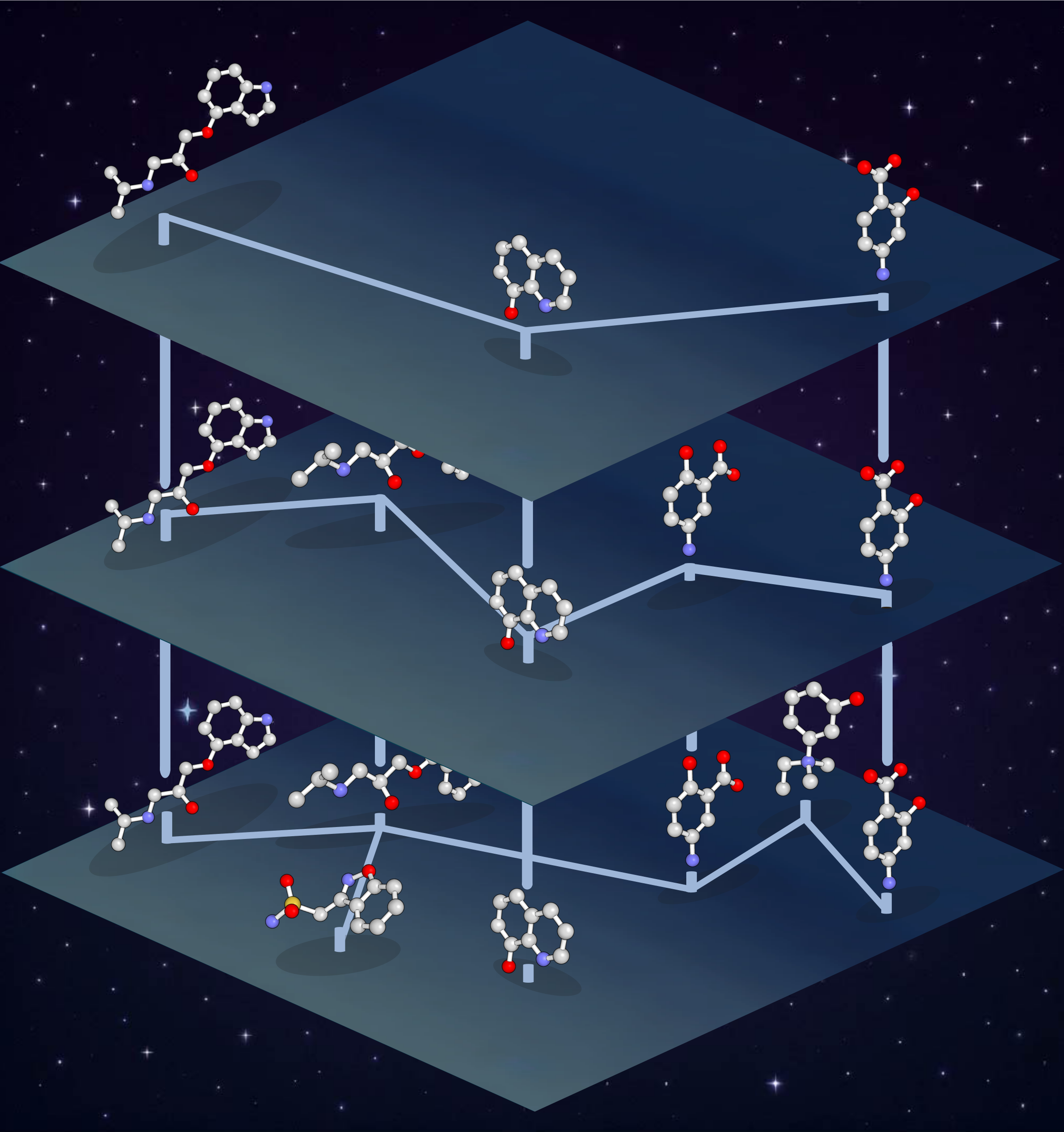
The RAD Method
Hierarchical Organization
Sparse hierarchy allows rapid identification of promising areas of chemical space
Small-World Graphs
Graph edges are based on Tanimoto similarity and chosen to maintain the small-world property, enabling efficient exploration
Best First Traversal
User-defined scoring functions guide graph traversal to quickly find good-scoring molecules while avoiding unpromising areas of chemical space
Distributed Scoring
Lightweight scoring workers coordinate with a single central process to enable massively parallel scoring
Why Choose RAD?
Efficient Screening
RAD can find over 50% of the top-scoring molecules in a library, while doing the expensive scoring calculation for only 1%.
Billion-Scale Validated
Proven performance on billion-scale chemical libraries with traditional molecular docking and modern ML models.
Distributed Computing
Scale from single machines to HPC clusters with Redis-coordinated workers that can run anywhere.
Flexible Infrastructure
Deploy locally, use our hosted public service to avoid setup complexity, or run your own infrastructure.
Get Started & Learn More
Installation & Documentation
Install
Publications
Original RAD Paper
Read the original paper describing the method and showcasing the performance on the DUDE-Z dataset.
Read PaperBillion-Scale Application
See RAD applied to billion-scale chemical libraries with UCSF DOCK and the ML model Chemprop.
Read Paper